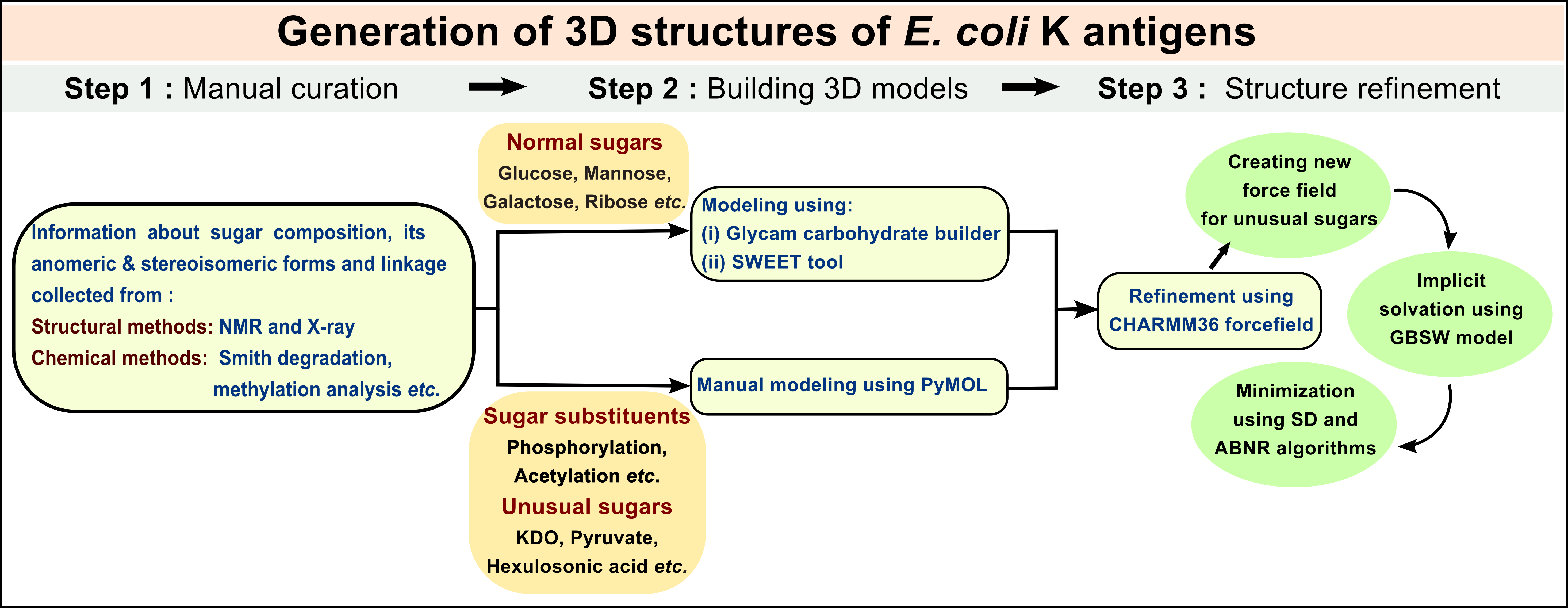
Generation of 3D structures of E. coli K antigens

Nomenclature used in chemical representation of monosaccharides in EK3D
|
OAc : O acetylation NAC : N acetylation Pyr : Pyruvate Glcp : Glucopyranose Galp : Galactopyranose Manp: Mannopyranose Quip: Quinovopyranose Fucp: Fucopyranose Rhap: Rhamnopyranose Galf: Galactofuranose Ribf: Ribofuranose Fruf: Fructofuranose GlcNAc : N Acetyl Glucosamine GalNAc : N Acetyl Galactosamine ManNAc : N Acetyl Mannosamine FucNAc : N Acetyl Fucosamine KDOp :KDO pyranose form (KDO : 3-Deoxy-D-manno-oct-2-ulosonic acid ) KDOf :KDO furanose form SugarP: 2,3-diacetamido-2,3,6-trideoxy-beta L-Mannopyranose SugarS: 6-O-Acetyl-4-deoxy-hexulosonic acid Gro : Glycerol Ser : Serine Thr : Threonine |
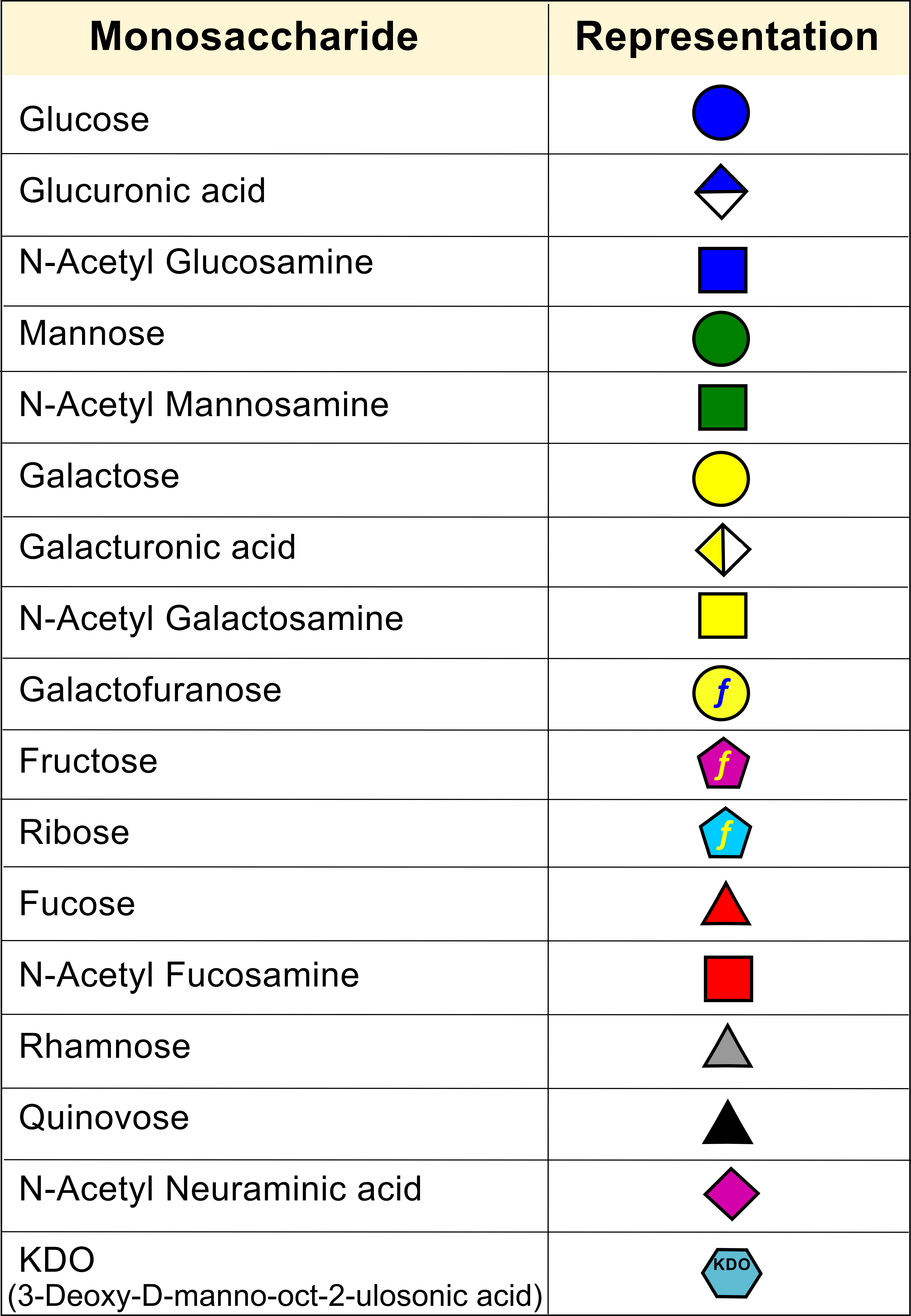
Schematic representation of monosaccharides in EK3D
|
1. Anomeric form (alpha/beta) is indicated at the top right of the sugar 2. Isomeric form (L/D) is indicated at the bottom left of the sugar 3. Furanose sugars are indicated by "f" inside the sugar (ex. Ribose in the table below) |
 |
The schematic representation has been adopted from the glycan representation developed by the Consortium for functional glycomics (CFG)
References:
- Berger, O., McBride, R., Razi, N. & Paulson, J. (2008) "Symbol Notation Extension for Pathogen Polysaccharides", The Scripps Research Institute, Consortium for Functional Glycomics.
- Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Marth, J.D., Bertozzi, C.R., Hart, G.W. and Etzler, M.E. (2009) Symbol nomenclature for glycan representation. Proteomics, 9, 5398-5399.
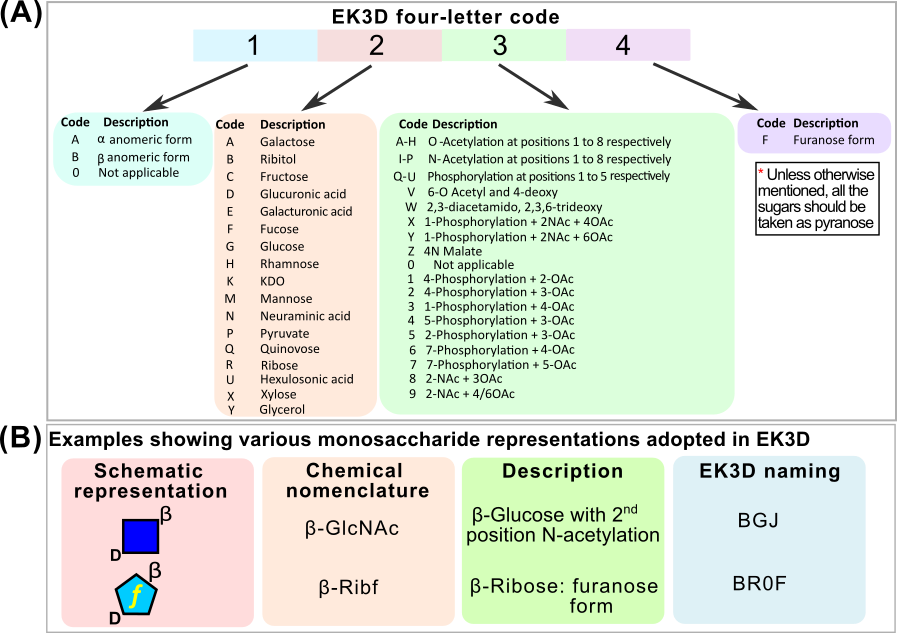
Four letter monosaccharide code used in EK3D